

Genetics and Plant Breeding
BIOTECHNOLOGY
RESEARCH ACHIEVEMENTS
TISSUE CULTURE
In vitro regeneration of coconut plantlets from immature inflorescence
Immature inflorescences of 2–12 cm size were collected from West Coast Tall variety and the rachilla segments were cultured on four different media combinations in dark condition. White translucent outgrowths were maximum in Y3 medium supplemented with 2, 4-dichlorophenoxyacetic acid. After eight weeks in dark, shoot-like outgrowth was noticed. After eight months dark incubation, multiple shoot formation was noticed in 1/2 MS with naphthalene acetic acid (NAA) and 6-benzylaminopurine (BAP). Maximum shoot development was observed Y3 medium fortified with 2-isopentenyl adenine (2ip) and BAP. The individual shoots after development of 3–4 leaves were transferred to 1/2 Y3 medium supplemented with NAA and μM indole-3-butyric acid (IBA) for root initiation. Start codon targeted (SCoT) profiling based on banding pattern of PCR-amplified products confirmed the clonal fidelity of in vitro regenerated coconut plantlets.

Arecanut immature inflorescence culture
A repeatable protocol for arecanut somatic embryogenesis and plantlet regeneration from immature inflorescence explants was developed at ICAR-Central Plantation Crops Research Institute, Kasaragod for rapid multiplication of elite genotypes such as Yellow leaf Disease (YLD) resistant palms, Hirehalli Dwarf (HD), a natural mutant with short stature and for dwarf hybrids, VTLAH–1 and VTLAH–2. For mass multiplication immature inflorescence rachillae were chopped into 1-2 cm bits size explants and inoculated in Y3 basal medium for somatic embryogenesis. Calli were induced on subsequent subculture from higher to lower auxin concentrations. Somatic embryo formation was achieved in hormone-free Y3 medium and germination was achieved when supplemented with cytokinins. Subsequent plantlet development was achieved in regeneration medium under light room condition with photoperiod of 16 hours. The tissue culture technology for mass multiplication of dwarf hybrids has also been transferred to a Tamil Nadu-based company (M/S SPIC Biotech Pvt. Ltd.), which in turn is likely to start its commercial seedling production from this year.

Immature inflorescence culture in arecanut dwarf hybrids.
Callus initiation, in vitro plantlets from somatic embryos and ex vitro hardened plantlet

Immature inflorescence culture in tall accession and YLD resistant arecanut palms
Protocol for in vitro propagation of coconut via plumule culture
A protocol has been standardized for regeneration of complete plantlets via organogenesis and embryogenesis from plumular tissues of West Coast Tall, Chowghat Green Dwarf, Malayan Orange Dwarf and Malayan Yellow Dwarf cultivars of coconut. Callus was induced from plumular tissues in Y3 media supplemented with either 2,4-D (74.6 μM) alone or 2, 4-D (74.6 μM) in combination with TDZ (4.54 uM). Higher percentages of embryogenic calli, somatic embryoids and meristemoids were obtained in Y3 media supplemented with either spermine (100 μΜ) or BA (4.44 μΜ).

Somatic embryogenesis and regeneration from plumular explants of coconut
EMBRYO CULTURE AND EMBRYO RESCUE
Coconut embryo culture
Production of planting material for propagation in coconut is solely through seed nuts since other vegetative means are not available. Field gene banks host genetic resources in coconut. The collection and transportation of coconut for the safe movement of coconut germplasm through embryo cultures, instead of seed nuts, is recommended by the technical guidelines finalized by FAO/IPGRI (currently Bioversity International). This method also avoids the formalities of quarantine regulations, which include treatments of the nuts with insecticide, fungicide and fumigants. Further, collection of embryos in vitro also greatly reduces the cost of transport. ICAR-CPCRI has developed a successful protocol for culturing coconut zygotic embryos from 8-11 months old nut, which was successfully utilized in the germ plasm expeditions of the institute. This technique makes use of effective artificial media with available macro and micronutrients and microclimatic conditions which support the embryo to grow and to form entire plantlets. The embryo culture protocol developed by ICAR-CPCRI was first implemented during 1994 for six Pacific Ocean accessions maintained at the World Coconut Germplasm Center, Andaman Islands wherein 87 embryos were brought to main land. Out of 83 plantlets retrieved, 25 plantlets were field planted at International Coconut Gene Bank for South Asia (ICG-SA) Kidu, Karnataka during 1996. Later five international expeditions were conducted by ICAR-CPCRI during the period 1997- 2001 for the collection of coconut genetic diversity. A total of 4182 embryos of 45 accessions were collected from eight countries, viz., Mauritius, Madagascar, Seychelles, Maldives, Comoros, Reunion, Sri Lanka and Bangladesh.

In vitro retrieval via embryo rescue of sweet endosperm coconut
MohachaoNaral is a variant in coconut with sweet and soft endosperm found in Ratnagiri district, Maharashtra, India. Efforts are made to raise tiny slender embryos of MohachaoNaral type which lack vigour and natural germination, in vitro. The plantlets resulted through in vitro cultured are successfully planted in the field.

Field collection and in vitro culturing of MohachaoNaral type coconut.
Harvesting of matured coconut (a); surface sterilization of endosperm plug with embryo (b); transportation of packed endosperm plug in ice box (c), embryo of MohachaoNaral type (d) in vitro germination of embryo (e-h); hardening in pot and poly bag (I and j) and field planting (k) of embryo cultured MohachaNaral type plantlet at Regional Coconut Research Station (RCRS), Bhatye, Ratnagiri, India.
CRYOPRESERVATION
Cryopreservation of coconut zygotic embryo
Cryopreservation technique is widely used for long-term conservation as it is safe and enables conservation of large genetic variability. Coconut pollen and embryo cryopreservation are an excellent option for long-term conservation of genetic resources, which can provide a viable backup to field gene banks. In case of embryos, pre treatment is very much essential before conserving in liquid nitrogen at -196°C. Air desiccation treatment resulted in maximum retrieval of healthy plantlets from the embryos subjected to 18 hours silica gel or 24 hours laminar air flow desiccation. Cryopreservation of mature coconut embryos through vitrification is suitable for the conservation. The PVS 3 solution was most effective for regeneration of cryopreserved embryos among the seven plant vitrification solutions tested. Most effective protocol involved preculture of embryos for three days on medium with 0.6 M sucrose, followed with PVS3 treatment for 16 h thereafter cooling rapidly in liquid nitrogen and rewarming and unloading in 1.2 M sucrose liquid medium for 1.5 h. The survival rates of 70-80 per cent (corresponding to size enlargement and weight gain) was achieved with this protocol and 20-25 per cent of the plants regenerated (showing normal shoot and root growth) from cryopreserved embryos could be established in pots and subsequently in the field.

Cryopreservation of coconut pollen
Coconut pollen is another excellent source of diverse alleles within a gene pool. Pollen cryopreservation has been successfully used in variety of plant species. Life span of most coconut pollen is for few days. Through pollen cryopreservation technology the pollen from desirable palms can be stored over a long period of time in addition to storage of multiple genotypes in small space for its future utility in case of threat from biotic and abiotic factors. The protocol developed by ICAR-CPCRI is proven to be simple and highly effective for the implementation of pollen cryobank to complement the field germplasm collections as well as for its utilization in future breeding programs. The methodology of pollen cryopreservation is briefly as follows: Extraction of pollen was done by sieving (mesh size - 0.2 mm) the male flowers which were incubated at 40°C for 24 h in an oven. Pollen were wrapped in aluminium foil strips and inserted into cryovials and plunged into the liquid nitrogen. Seed set after hand pollination using cryopreserved pollen for a period of four years was found to be normal one hundred percent germination was observed in embryos extracted from hybrid nuts produced with cryostored pollen and plantlet development was normal

Cryopreservatory of coconut pollen for utilization in resistance breeding program
The demand for seedlings of KalpaSankara (CGD X WCT) hybrid notified and released for the root (wilt) disease prevalent tract is very high. In order to enhance the hybrid seedling production, high yielding disease-free Chowghat Green Dwarf (CGD) mother palms located in farmers’ plot are selected for artificial pollination to be carried out in a decentralized mode. The male flowers are collected from ELISA tested root (wilt) disease-free West Coast Tall (WCT) parental palms and processed at ICAR-CPCRI, Regional Station, Kayamkulam and stored in deep freezer/ liquid nitrogen. The stored WCT pollen will be transported to each Panchayat/KrishiBhavan and kept in desiccators. Through this centralized mechanism of pollen processing, disease-free status of the male parental palms and quality of the pollen will be assured. Presently 60 high yielding and disease-free WCT male parental palms are used for collecting male flowers. From one inflorescence, approximately 6-8 g of pollen is obtained, which is transferred to 8-10 cryo vials (2.0 ml capacity). Cryo vials are stored in liquid nitrogen cans; a can of 121 litre capacity can store up to 6000 samples. Pollen vials are taken out from the cans as per requirement and one vial (filled with 1.5 gram pollen) can be used for pollinating six CGD palms for two days.
Arecanut pollen collection and cryopreservation
Various aspects of arecanut pollen cryopreservation viz. desiccation and collection of pollen, in vitro germination, viability and fecundity studies has been standardized.In vitro viability tests were conducted using fresh and desiccated pollen of three arecanut genotypes (Sumangala, Hirehalli Dwarf and Hirehalli Dwarf x Sumangala). The desiccated pollen was cryopreserved by direct immersion in liquid nitrogen for different durations (24 hours to two years). Viability and fertility studies were conducted using cryopreserved pollen. Pollen extraction was achieved from fully opened male flowers by desiccation at room temperature (33- 34ºC). Sucrose at a concentration of 2.5 g/l was found to be best for in vitro germination at room temperature among different sucrose media tested. There was no significant difference between desiccated and cryopreserved pollen for germination, but pollen tube length decreased significantly in cryopreserved pollen in Hirehalli Dwarf and Sumangala. Fertility studies using cryopreserved arecanut pollen from HD x Sumangala at different duration’s viz., one month, one year and two years showed setting percentages of 70, 43 and 62, respectively. Normal nut set was observed with the use of cryopreserved pollen

Arecanut pollen cryopreservation
a) Fully opened inflorescence of Hirehalli Dwarf arecanut palm
b) Magnified view of opened male flowers
c) Magnified view of dehisced anthers
d) Collection of pollen
e) Germination of fresh pollen
f) Germination of desiccated pollen
g) Germination of cryostored pollen
h) Receptive stigma
(i) Nut set using pollen cryopreserved for 24 h
(j) Two years; k) Seedling raised from nut obtained through pollination using two years old cryopreserved pollen
Cryopreservation of arecanut embryogenic calli
Arecanut embryogenic calli were pre-grown in Y3 medium supplemented with sucrose (0.2, 0.3 and 0.4 M) for three days and later desiccated using plant vitrification solution 3 (PVS3) for 30 minutes. The results showed 8-10 % recovery of embryogenic calli that resulted in normal plantlet production. The clonal fidelity studies, using SCoT marker, showed no variation of cryopreserved calli in comparison to the original calli. This preliminary study demonstrated a successful use of V-cryoplate technique in cryopreservation of embryogenic calli of arecanut. With better recovery percentage, the optimal concentration of sucrose in the pre-culture medium was found to be 0.3 M. Vitrification in PVS3 solution for 30 minutes had no adverse effect. With this technique it is possible to store the embryogenic potential of arecanut calli for long term.

V-cryoplate cryopreservation of embryogenic calli in arecanut
(a) Arecanut embryogenic calli generated from immature inflorescence culture
(b) fixing the embryogenic calli explants on V-cryoplate
(c) V-cryoplates accommodated completely with the explants
(d) encapsulation using sodium alginate (3%) and calcium chloride (0.01 M)
(e) dehydration in PVS3 solution (V-cryoplate)
(f) plunging the cryovials containing cryoplates in liquid nitrogen.
Coconut pollen collection and cryopreservation during monsoon season
It is feasible to collect coconut pollen during monsoon season by slow drying initially in room temperature and subsequently at 35ºC for 7 hrs followed by 40ºC for 12 hrs. The retention of viability of pollen collected using this method even after cryopreservation would be beneficial for its use in breeding programmes as well as for the production of hybrids which are in high demands. This study thus paves the way for collection of pollen resources during wet season and cryostoring them for future use.

Normal nut set in West Coast Tall palm pollinated with pollen collected and cryopreserved during monsoon season
Molecular-marker based characterization of coconut germplasm and genetic diversity assessment
Microsatellite-based characterization of coconut accessions from Andaman and Nicobar Islands (100 palms, 26 accessions, Amini and Kadmat islands (56 palms, 9 accessions) and Minicoy island (67 palms, 4 accessions) of Lakshadweep, tall germplasm (243 palms, 20 accessions), dwarf germplasm (134 palms, 24 accessions), ‘MohachaoNarel’, a sweet kernel type from Maharashtra (28 palms) was carried out and the population structure and extent of genetic diversity within these accessions were deduced. Characterization of Annur, Bedakam and Kuttiyadi ecotypes and comparison of its similarity/diversity with West Coast Tall (WCT) cultivar was also carried out using microsatellite markers. Data analysis revealed that three ecotypes were distinct compared to the WCT population, from which they have been assumed to have arisen
PCR amplification pattern of microsatellite loci CNZ4 with sweet kernel coconut genotypes (‘MohachaoNarel’)

M: Molecular weight ladder; Lanes 1-26: Sweet kernel coconut population; Lanes 27-29: Control palms
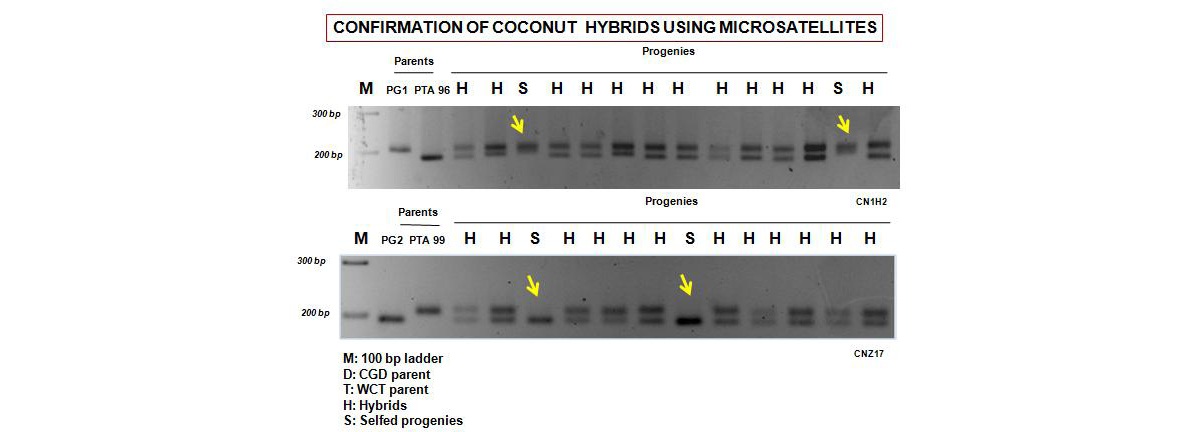
Development of microsatellite panel for hybrid purity assessment in coconut
It is difficult to identify genuine hybrids in the nursery stage relying solely on morphological markers. A molecular technique to assess the genetic purity of coconut hybrids (Dwarf x Tall) has been developed using SSR (Simple Sequence Repeats) markers. A panel of 17 informative SSR markers was identified, which were capable of distinguishing parental lines, viz., Chowghat Green Dwarf (CGD) and West Coast Tall (WCT) used for hybrid (DxT) coconut production. These markers were utilized in DxT hybrid seedling purity assessments in coconut nurseries. Off-type seedlings (selfed ones) could easily be distinguished from hybrids using these markers and eliminated before distribution to farmers
Start codon targeted polymorphism (SCoT) markers were utilized for assessment of genetic diversity in 23 coconut accessions (10 talls and 13 dwarfs), representing different geographical regions. A total of 15 SCoT primers were selected based on their consistent amplification patterns. A total of 102 scorable bands were produced by the 15 primers, 88 % of which were polymorphic. The scored data were used to construct a similarity matrix. The similarity coefficient values ranged between 0.37 and 0.91. These coefficients were utilized to construct a dendrogram using the unweighted pair group of arithmetic means (UPGMA). The extent of genetic diversity observed based on SCoT analysis of coconut accessions was comparable to other marker systems. Tall and dwarf coconut accessions were clearly demarcated, and in general, coconut accessions from the same geographical region clustered together.
Start codon targeted (SCoT) markers for assessment of genetic diversity of arecanut
Analysis of genetic diversity among six arecanut accessions, viz., Mangala, Sumangala, Sreemangala, Mohitnagar, Swarnamangala and a natural dwarf mutant (Hirehalli Dwarf) was carried out using SCoT markers. Using 10 SCoT primers, 82 band were produced among the accessions, of which 58 (70.73%) were found to be polymorphic. The highest genetic similarity value of 0.89 was found between the Swarnamangala and Mohitnagar and the lowest value of 0.63 was noticed between the Hirehalli Dwarf and Mohitnagar. The similarity coefficient values were then utilized to construct a dendrogram utilizing the unweighted pair group of arithmetic means (UPGMA). The cultivars were grouped depending on their geographical origins.
SCoT banding pattern of arecanut accessions with SCoT11 primer
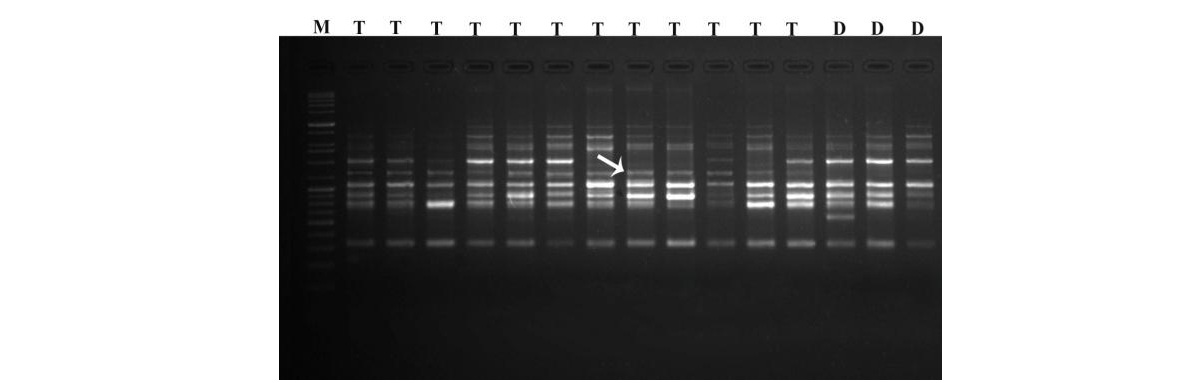
T: Tall accessions (Mangala, Sumangala, Sreemangala, Mohitnagar, Swarnamangala and Hirehalli Tall) and D: dwarf palm (Hirehalli Dwarf )
Identification of molecular markers linked to tall/dwarf trait in coconut
RAPD markers for tall/dwarf-type palm traits were identified and sequence characterized amplified region (SCAR) primers were designed from the unique RAPD amplicons. The SCAR primers were validated in coconut cultivars representing different geographical regions. The identified SCAR markers are presently being utilized for juvenile selection of dwarf/tall coconut palms in the nursery stage, which was earlier carried out by using morphological markers (which are unreliable).
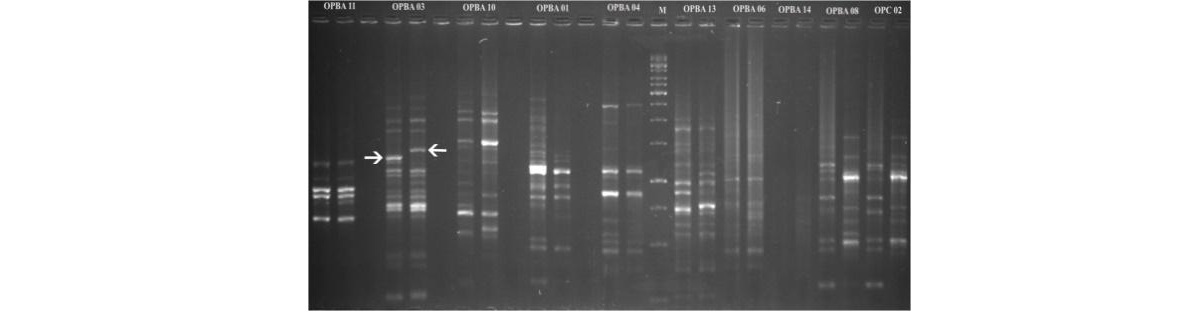
RAPD banding patterns of coconut tall and dwarf bulks with different primers
Arrowheads indicate polymorphic bands of OPBA03 (arrow heads) distinguishing tall and dwarf pooled DNA.
Development of a technique for hybrid detection in coconut using EST-SSRs
Thirty EST-SSR primers were used to screen for polymorphism between 18 coconut parental lines and selected polymorphic primers were used for authentication of hybrids in 103 progenies derived from the 18 crosses. True hybrids possessed banding pattern of both parental lines. This technique can enable identification of true hybrid seedlings in the nursery stage itself, before distribution to the farming community.
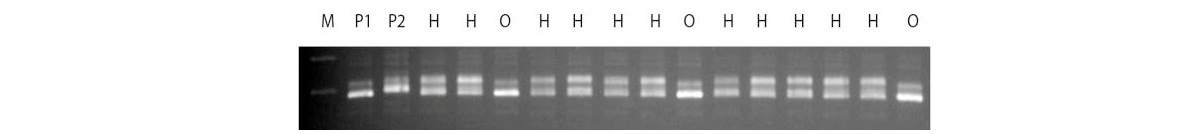
Gel profile of coconut parents [Chowghat Green Dwarf (CGD) x West Coast Tall (WCT)] and their hybrids. M: 100bp ladder, P1: Female parent (CGD),
P2: Male parent (WCT), H: Hybrids, O: Off-types
Arecanut cultivars are generally tall, which is a major constraint in arecanut cultivation. High yielding tall cultivars have been crossed with natural dwarf mutant, Hirehalli Dwarf (HD) and hybrids have been developed and released for commercial cultivation. It is very difficult to distinguish selfed progenies of the female mother palm in the nursery stage using morphological traits. Start Codon Targeted (SCoT) primers were used to screen tall cultivars of arecanut and HD. One of the primers, SCoT11, produced an amplicon of around 1300 bp band, which was present in all the tall cultivars, but was absent in the dwarf accession. A SCAR marker, capable of distinguishing tall/dwarf trait in arecanut, was also developed and validated, which could ensure supply of genuine hybrid planting material to the farming community.
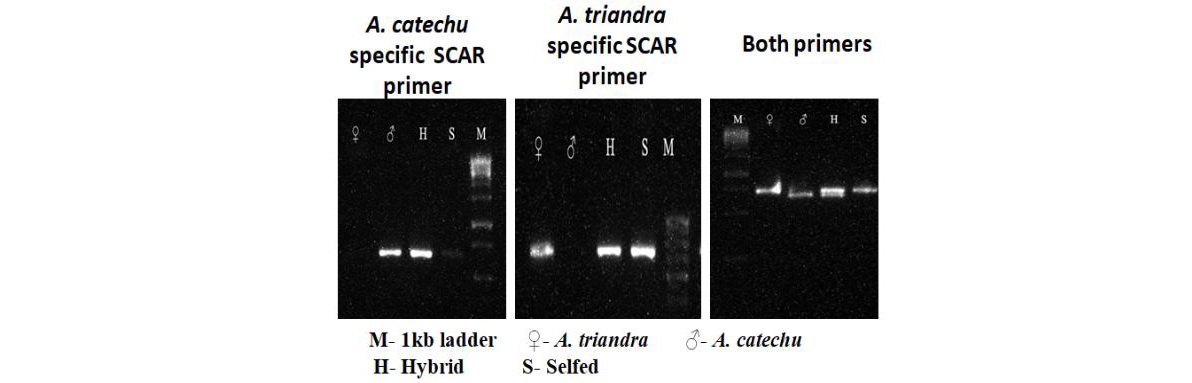
Development of SCAR markers for detection of interspecific hybrids in arecanut
Areca catechu and its wild relativesviz., Areca concinna and Areca triandra were characterized using Start Codon Targeted polymorphism (SCoT). Markers unique to A. catechu and A. triandrawere identified. Based on these sequences, SCAR markers specific to A. catechu and also to A. triandra were designed. Authentic inter-specific hybrids between Areca catechu and Areca triandra could be identified utilizing these SCAR markers.
Microsatellite-based characterization of cocoa germplasmand genetic diversity assessment
Eleven genotypes of cocoa from South and Central America viz., Columbia (SC-1, SC-9, SC-4), Brazil (RB-33/3), Peru (POU- 16/A, POU-7/B), Costa Rica (RIM-41, RIM-189) and Ecuador (B-12/2, JA-1/19, AMAZ-15) and 44 Nigerian accessions have been characterized using highly polymorphic SSR primer pairs. Genetic diverse parents have been identified which are presently being utilized in cocoa breeding programmes for exploiting heterosis.
Polymorphic banding patterns of cocoa accessions from South and Central America
Identification of a molecular marker linked to sex determination in palmyrah
Palmyrah is a dioecious palm, with female trees being economically important. The palms are slow growing and have no distinguishing features to determine sex until flowering, which commences 12-15 years of maturity. Breeding would be facilitated if gender could be identified at seedling stage itself. RAPD primer OPA-06 produced a 600 bp marker only in male palms and is useful for sex determination in palmyrah.
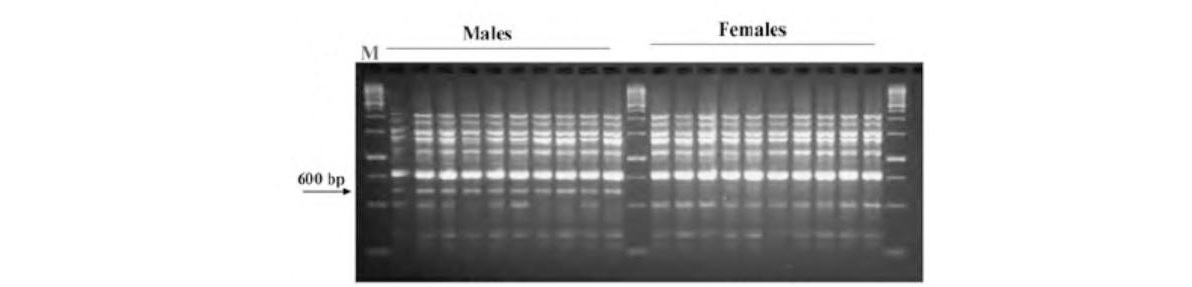
RAPD banding pattern of male and female palmyrah palms usingRAPD primer OPA-06. Arrow indicates 600 bp male-specific band.
GENOMICS, TRANSCRIPTOMICS AND PROTEOMICS
Genome assembly and annotation of the dwarf coconut (Cocos nucifera L. cv. Chowghat Green Dwarf)
The genome of the dwarf coconut cultivar ‘Chowghat Green Dwarf’ (CGD) of 1.93 Gb, was assembled using different paired-end and mate-pair Illumina reads and long reads from PacBio single molecule sequencing. Around 77 % of the draft genome is comprised of transposable elements and repeats. We predicted 51,953 protein-coding genes in the draft genome of which 47,746 (91.9%) could be annotated. Phylogenetic analysis based on single-copy genes and one-to-one orthologous genes placed the dwarf coconut close to oil palm and date palm. Further, the CGD genome shared substantial syntenywith the oil palm genome. The availability of dwarf coconut genome offers coconut researchers and breeders a means for deducing the origin of dwarf coconut cultivars, dissection of genes controlling plant habit and fruit color and accelerated breeding for improved agronomic traits.
Identification of expressed resistance gene analogue (RGA) sequences in coconut leaf transcriptome
Comprehensive bioinformatics analysis of RNA-Seq dataset of leaf of coconut root (wilt) disease-resistant cultivar Chowghat Green Dwarf resulted in the identification of 243 resistance gene analog (RGA) sequences, comprising six classes of RGAs. Domain and conserved motif predictions of clusters were performed to analyze the architectural diversity. Phylogenetic analysis of deduced amino acid sequences revealed that coconut NBS-LRR type RGAs were classified into distinct groups based on the presence of TIR or CC motifs in the N-terminal regions.
Global transcriptome profiling of leaves of healthy and diseased Chowghat Green Dwarf (CGD) palms was conducted to explore the molecular mechanisms involved in compatible and incompatible interactions,. RNA-Seq analysis generated more than 190 million 100 bp reads from both healthy and diseased samples. Assembled transcriptome yielded 59,282 transcripts with a median length of 987 bp. BLASTX annotation of transcriptome resulted in 39,665 transcripts getting annotated from Uniprot and date palm proteome database. Differential gene expression profiling analysis revealed 2718 transcripts to be up- or down- regulated in the diseased palms in comparison to healthy control at a fold change of 2 and above with a p value <=0.05. The differentially expressed transcripts could be categorized into pathways which included cell wall biogenesis, primary and secondary metabolism, plant-pathogen interaction, cellular transport, hormone biosynthesis and signaling.
Model for coconut defense response to root (wilt) disease
De novo assembly and characterization of global transcriptome of embryogenic calli in coconut
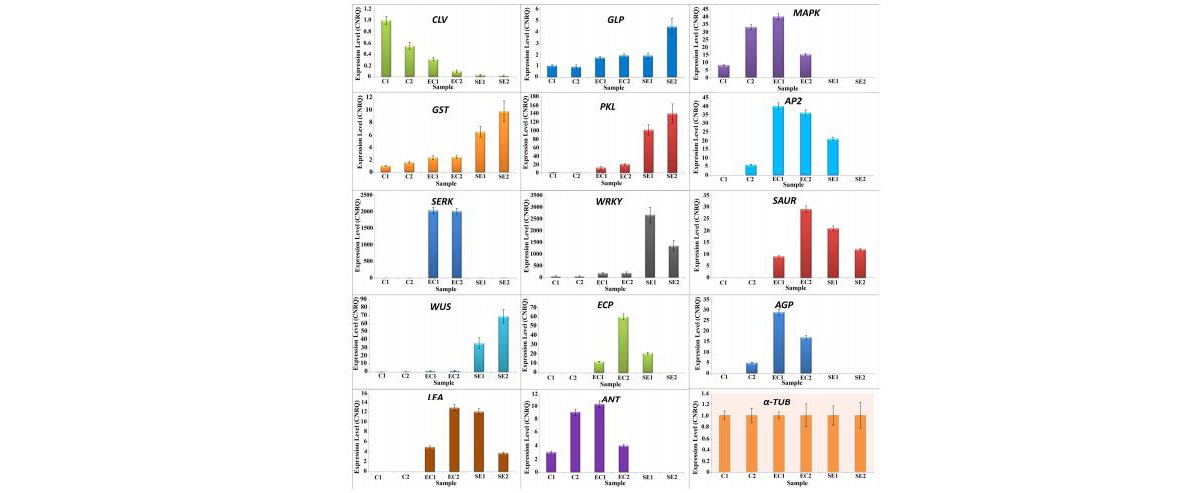
Transcriptome analysis (RNA-Seq) of coconut embryogenic calli, derived from plumular explants of West Coast Tall cultivar, was undertaken on an Illumina HiSeq 2000 platform. After de novo transcriptome assembly and functional annotation, a total of 40,367 transcripts were obtained, which showed significant BLASTx matches with similarity greater than 40 % and E value of ≤10−5. Fourteen genes known to be involved in somatic embryogenesis were identified. Quantitative real-time PCR (qRT-PCR) analyses of these 14 genes were carried in six developmental stages. The result showed that CLV was upregulated in the initial stage of callogenesis. Transcripts GLP, GST, PKL, WUS, and WRKY were expressed more in somatic embryo stage. The expression of SERK, MAPK, AP2, SAUR, ECP, AGP, LEA, and ANT were higher in the embryogenic callus stage compared to initial culture and somatic embryo stages.
Calibrated and normalized relative gene expression (CNRQ) levels of 14 transcripts of coconut plumular-derived callus, along with the endogenous control, alpha-tubulin (α-TUB)
Differential expression pattern of microRNAs (miRNAs) and associated target genes during coconut somatic embryogenesis
Genome-wide profiling of small RNAs from embryogenic (EC) and non-embryogenic calli (NEC) was undertaken using Illumina Hiseq 2000 platform. A total of 110 conserved miRNAs (representing 46 known miRNA families) in both types of calli were identifie. In addition, 97 novel miRNAs (48 specific to EC, 21 specific to NEC and 28 common to both the libraries) were also identified. Among the conserved miRNAs, 10 were found to be differentially expressed between NEC and EC libraries with a log2 fold change > 2 following RPM-based normalization. miR156f, miR167c, miR169a, miR319a, miR535a, and miR5179 are upregulated and miR160a, miR166a, miR171a, and miR319b are down-regulated in NEC. To confirm the differential expression pattern and their regulatory role in SE, the expression patterns of miRNAs and their putative targets were analyzed using qRT- PCR and most of the analyzed miRNA-target pairs showed inverse correlation during somatic embryogenesis. Selected targets were further validated by RNA ligase mediated rapid amplification of 5′ cDNA ends (5′RLM-RACE).
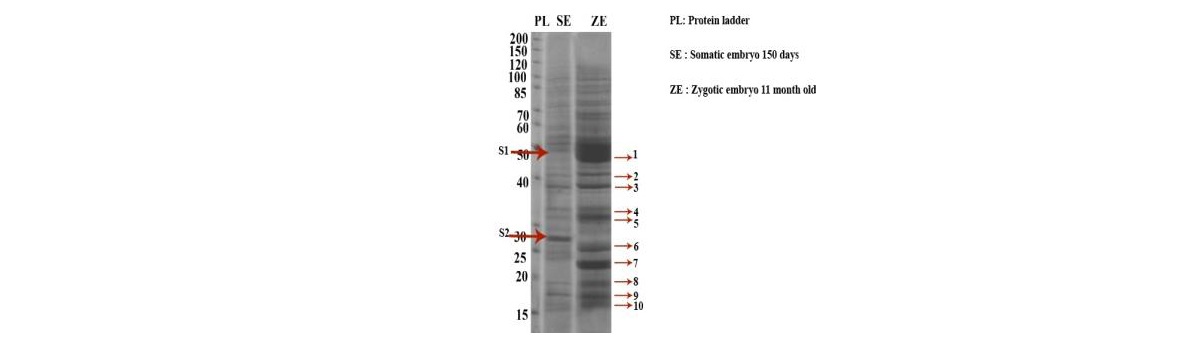
Identification of differentially expressed proteins in various stages of zygotic and somatic embryogenesis
A comparative study at proteomic level Was conducted between the different stages of coconut somatic embryos and zygotic embryos. The peptides differentially expressed were characterized by matrix assisted laser desorption ionization time-of-flight tandem mass spectrometry (MALDI-TOF/TOF MS) acquisition in an proteomics analyzer.
Comparative analyses of proteins expressed during coconut zygotic and somatic embryogenesis
NANOTECHNOLOGY
Coconut inflorescence sap mediated synthesis of silver nanoparticles and its antimicrobial properties
Coconut inflorescence sap was used to synthesize silver NPs (AgNPs). Metabolomic profiling of coconut inflorescence sap from West Coast Tall cultivar was initially undertaken to delineate its individual components. It was found to comprise of 64% secondary metabolites, 9% sugars, 12% lipids/fats and 9% peptides in positive mode, whereas in the negative mode, it was 33, 20, 9 and 11%, respectively. The concentration of silver nitrate, inflorescence sap and incubation temperature for the synthesis of AgNPs were optimized. Incubating the reaction mixture at 40 °C was found to enhance AgNP synthesis. The AgNPs synthesized were characterized using UV–visible (UV-Vis) spectrophotometry, X-Ray Diffraction (XRD), Fourier Transform Infrared spectroscopy (FTIR) and Transmission Electron Microscopy (TEM). The particles were crystalline in nature and the bulk of the particles were spherical with smooth (thin) shell and poly-dispersed with a diameter ranging from 10 nm to 30 nm. Antimicrobial property of AgNPs was tested in tissue culture of arecanut (Areca catechu L.) where bacterial contamination (Bacillus pumilus) was a frequent occurrence. A significant reduction in the contamination was observed when plantlets were treated with aqueous solutions of AgNPs. Notably, treatment with AgNPs did not affect the growth and development of the arecanut plantlets.
BIOINFORMATICS
Refinement of protocol for in vitro regeneration in coconut
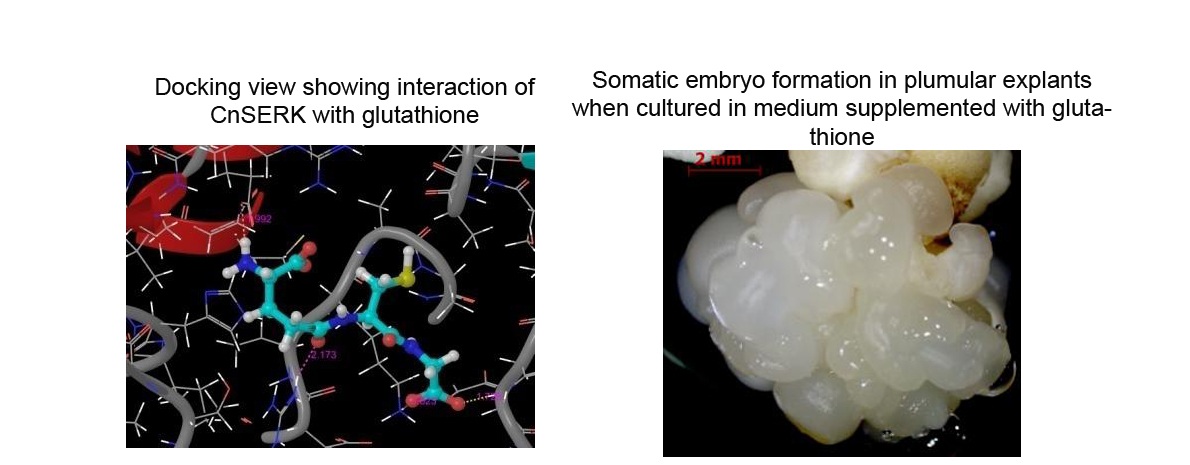
Homology–based modeling and Molecular Dynamics (MD) simulation of a coconut somatic embryogenesis receptor-like kinase protein (CnSERK) was carried out for exploring its structural features, functional characterization of its active sites and binding mechanisms of selected plant hormones and growth regulators by docking studies. In vitro studies were carried out to compare the efficiency of three selected chemicals [adenine sulphate, glutathione and 22(S), 23(S)-homobrassinolide] in enhancing somatic embryogenesis from plumular explants of coconut; glutathione gave the best response for induction of both embryogenic calli and somatic embryogenesis.
Sequences representing resistance gene analogues (RGAs) of coconut, arecanut, oil palm and date palm were collected from NCBI, sorted based on domains and assembled into a database. The sequences were analyzed in PRINTS database to find out the conserved domains and their motifs present in the RGAs. Based on these domains, we have also developed a tool to predict the domains of palm R-genes using Support Vector Machines (SVM), stand alone BLAST and Hidden Markov Models (HMM).. The model files were selected based on the performance of the best classifier in training and testing. All these information is stored and made available in the online ‘PRGpred’database and prediction tool.
Availability:http://192.99.220.223/PalmRgene/
COCONUT EST-SSR DATABASE
A dynamic EST-SSR database, based on SSRs generated from coconut transcriptome datahas been developed, which is an excellent resource of novel markers for the coconut researchers and breeding community. This database has been so designed to provide a user-friendly, comprehensive web based resource detailing the information on EST-SSRs, generated through this study, and which potentially possess a wide range of genetic analysis and breeding applications.

Screen shot of home page of coconut
EST-SSR database
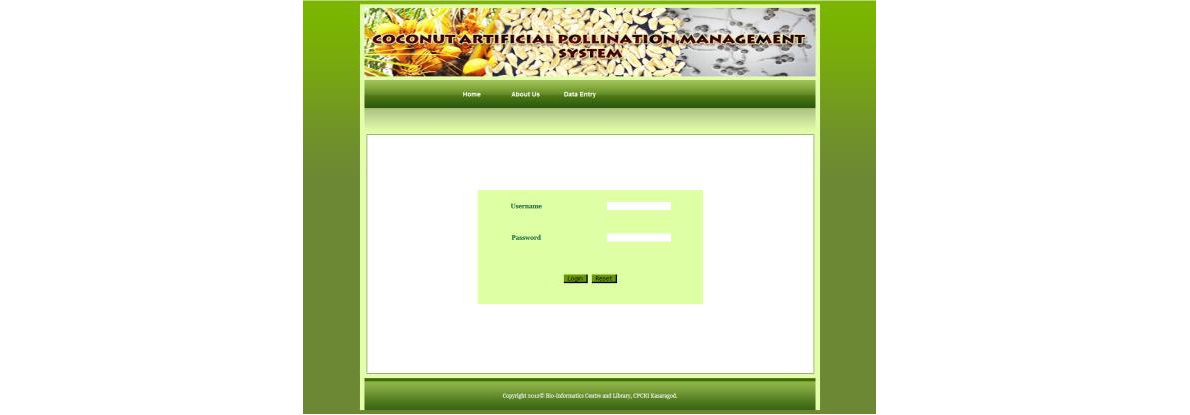
‘COCONUT ARTIFICIAL POLLINATION MANAGEMENT SYSTEM’
This expert system developed helps in decision making and tracking the process of artificial pollination. This database meets two requirements, review of work done and planning of future work. It also helps in supervision and monitoring the pollinators which are very important in timely completion of artificial pollination. For database implementation, data was stored in MySQL running on a Linux server. The database interface was implemented using PHP and HTML.
‘PEST AND DISEASE SURVEILLANCE DATABASE’
The web-based tool on pest and disease surveillance is designed to enable to understand the population dynamics of insect pests and disease incidence. Monthly incidence of pests and disease on coconut in CPCRI experimental farm is uploaded. This data base also provides information on pests and diseases in coconut and its management.
Availability:http://14.139.158.118/bioinfo_db/pest/
‘MAPS’: A TOOL FOR DETECTION OF MICROSATELLITES IN WHOLE GENOME SEQUENCES
MAPS (Microsatellite Analysis and Prediction Software), a bio-Java based independent; stand alone platform was designed to allow the identification and characterization of microsatellites in entire genomes. The algorithm is coded by JSP as the front end and MySQL as the back-end. The algorithm can extract perfect, imperfect and compound microsatellites from genome accurately and also, from coding and non-coding regions.
Availability:http://14.139.158.118:8080/MSQL/Home.jsp
‘PROMIT’: A tool for computational prediction of 70 promoters inPseudomonas spp.
An algorithm was developed for prediction of promoters in -10 and -35 regions of Pseudomonas spp. Promoter regions were predicted using both Support Vector Machines and Hidden Markov Model-based approaches.
SVM-based algorithm for prediction of antibiotic biosynthetic genes in plant growth promoting rhizobacteria
A method for prediction of enzymes involved in antibiotic biosynthetic pathways (phenazine, 2,4–diacetylphloroglucinol, pyrrolnitrin and pyoluteorin) in plant growth promoting Pseudomonas species on the basis of amino acid composition and di-peptide composition was developed by using the Support Vector Machines.
‘CMSD’: Database of microsatellite markers genotyped in indigenous coconut accessions
The increased use of molecular markers in varietal identification has led to the need for an appropriate storage place and this has paved way for the development of a marker database containing allelic data of coconut accessions as revealed by microsatellite markers. The main features of the information system are a SSR database, SSR analysis tool, variety identification tool, data submission form and brief information about microsatellites.
CnMAPKPred: A machine learning approach for predicting mitogen-activated protein kinase (MAPK)
A tool has been developed for the prediction of MAPK in coconut using Naïve Bayes algorithm.
BACPred: A tool for prediction of bacilysin synthesizing genes in Bacillus spp.
The biosynthetic genes involved in bacilysin production are bacABCDEoperon and a monocistronic gene ywfH. Using machine learning algorithm, WEKA, a tool was developed to predict these bacilysin synthesizing genes from whole genome of plant growth promoting Bacillus spp.
Screen shot of home page of BACPred
PHZ Pred – A TOOL FOR PREDICTION OF PHENAZINE SYNTHESIZING GENES
IN Pseudomonas spp.
Using machine learning algorithm, WEKA, a tool was developed to predict phenazinesynthesizinggenes (seven clusters; phzA to phzG) in genomes of plant growth promoting Pseudomonas spp.AwebinterfacewasdevelopedforthetoolusingHTML,PHPandJavaScript.
LTTRPRED: A TOOL FOR IDENTIFYING AND PREDICTING THE LTTR GENES
LTTRPred is a tool developed for identifying and predicting the LTTR genes, responsible for the activation of Plttranscription regulators, in whole genome of various Pseudomonas spp. LTTRPred was developed using support vector machine (SVM) based on the composition of amino acid and amino acid pairs.